R has some great functions for generating scatterplots in 3 dimensions. Two of the best are the scatter3d() function in John Fox’s car package, and the scatterplot3d() function in Uwe Ligges’ scatterplot3d package. In this post, we will focus on the later.
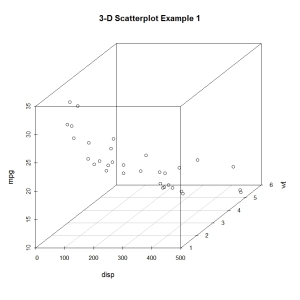
Let’s say that we want to plot automobile mileage vs. engine displacement vs. car weight using the data in the mtcars dataframe.
library(scatterplot3d)
with(mtcars, {
scatterplot3d(disp, # x axis
wt, # y axis
mpg, # z axis
main="3-D Scatterplot Example 1")
})
The resulting plot is given below.
Now lets, modify the graph by replacing the points with filled blue circles, add drop lines to the x-y plane, and create more meaningful labels.
library(scatterplot3d)
with(mtcars, {
scatterplot3d(disp, wt, mpg, # x y and z axis
color="blue", pch=19, # filled blue circles
type="h", # lines to the horizontal plane
main="3-D Scatterplot Example 2",
xlab="Displacement (cu. in.)",
ylab="Weight (lb/1000)",
zlab="Miles/(US) Gallon")
})
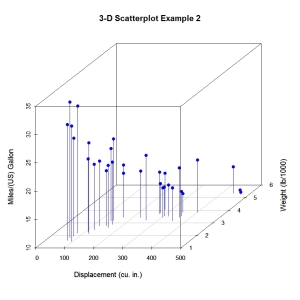
Next, let’s label the points. We can do this by saving the results of the scatterplot3d() function to an object, using the xyz.convert() function to convert coordinates from 3D (x, y, z) to 2D-projections (x, y), and apply the text() function to add labels to the graph.
library(scatterplot3d)
with(mtcars, {
s3d <- scatterplot3d(disp, wt, mpg, # x y and z axis
color="blue", pch=19, # filled blue circles
type="h", # vertical lines to the x-y plane
main="3-D Scatterplot Example 3",
xlab="Displacement (cu. in.)",
ylab="Weight (lb/1000)",
zlab="Miles/(US) Gallon")
s3d.coords <- s3d$xyz.convert(disp, wt, mpg) # convert 3D coords to 2D projection
text(s3d.coords$x, s3d.coords$y, # x and y coordinates
labels=row.names(mtcars), # text to plot
cex=.5, pos=4) # shrink text 50% and place to right of points)
})
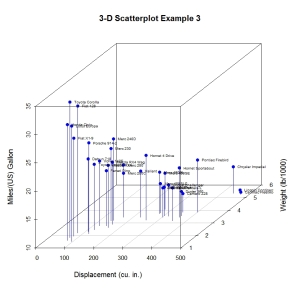
Almost there. As a final step, we will add information on the number of cylinders each car has. To do this, we will add a column to the mtcars dataframe indicating the color for each point. For good measure, we will shorten the y axis, change the drop lines to dashed lines, and add a legend.
library(scatterplot3d)
# create column indicating point color
mtcars$pcolor[mtcars$cyl==4] <- "red"
mtcars$pcolor[mtcars$cyl==6] <- "blue"
mtcars$pcolor[mtcars$cyl==8] <- "darkgreen"
with(mtcars, {
s3d <- scatterplot3d(disp, wt, mpg, # x y and z axis
color=pcolor, pch=19, # circle color indicates no. of cylinders
type="h", lty.hplot=2, # lines to the horizontal plane
scale.y=.75, # scale y axis (reduce by 25%)
main="3-D Scatterplot Example 4",
xlab="Displacement (cu. in.)",
ylab="Weight (lb/1000)",
zlab="Miles/(US) Gallon")
s3d.coords <- s3d$xyz.convert(disp, wt, mpg)
text(s3d.coords$x, s3d.coords$y, # x and y coordinates
labels=row.names(mtcars), # text to plot
pos=4, cex=.5) # shrink text 50% and place to right of points)
# add the legend
legend("topleft", inset=.05, # location and inset
bty="n", cex=.5, # suppress legend box, shrink text 50%
title="Number of Cylinders",
c("4", "6", "8"), fill=c("red", "blue", "darkgreen"))
})
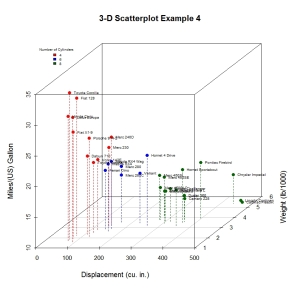
One of R‘s most attractive features is that it allows us to manipulate output and deeply customize graphs. This article has just touched the surface. Since colors and text labels can be input as vectors, you could programmatically use them to represent almost anything. For example, point colors and/or labels could be used to highlight observations that are outliers, have high leverage, or are unusual in some other way. Simply create a vector that has colors or labels for notable observations and missing (NA) values otherwise.