Permuation tests (also called randomization or re-randomization tests) have been around for a long time, but it took the advent of high-speed computers to make them practically available. They can be particularly useful when your data are sampled from unkown distributions, when sample sizes are small, or when outliers are present.
R has two powerful packages for permutation tests – the coin package and the lmPerm package. In this post, we will take a look at the later.
The lmPerm package provides permutation tests for linear models and is particularly easy to impliment. You can use it for all manner of ANOVA/ANCOVA designs, as well as simple, polynomial, and multiple regression. Simply use lmp() and aovp() where you would have used lm() and aov().
Example
Consider the following analysis of covariance senario. Seventy five pregnant mice are divided into four groups and each group receives a different drug dosage (0, 5, 50, or 500) during pregnancy. Does the dosage of the drug affect the birthweight of the resulting litters, after controlling for gestation time and litter size?
The data are contained in the litter dataframe available in the multcomp package. The dependent variable is weight (average post-birth weights for each litter). The independent variable is dose, and gestation time and litter size are covariates contained in the variables gesttime and number respectively.
If we were going to carry out a traditional ANCOVA on this data, it would look something like this:
> library(multcomp)
> data(litter)
> mod1 <- aov(weight ~ number + gesttime + dose, data=litter)
> summary(mod1)
Df Sum Sq Mean Sq F value Pr(>F)
number 1 69.8 69.77 4.367 0.04038 *
gesttime 1 143.7 143.70 8.994 0.00378 **
dose 3 122.8 40.93 2.562 0.06196 .
Residuals 68 1086.4 15.98
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
It appears that while litter size and gestation time are significantly related to average birthweight for a litter, the drug dosage is not (p = 0.062). (Note that the order of effects in the aov statement is important. Effects later in the list are adjusted for effects earlier in the list. This is the sequential or Type I sums of squares approach.)
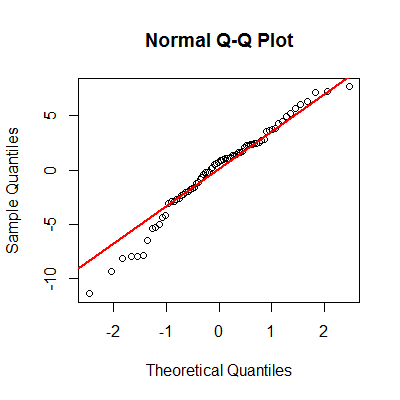
One of the assumptions of the ANCOVA model is that residuals are normally distributed. Let’s take a look.
> qqnorm(resid(mod1), main="Normal Q-Q Plot") > qqline(resid(mod1), col="red"))
From the graph, we have to question the normality assumption here. Note the deviations from the line.
An alternative to the tradional analysis of covariance is a permutation verion of the test. The test is valid even if we violate the normality assumption. To perform the test, simply replace the aov() function with aovp().
> library(lmPerm)
> mod2 <- aovp(weight ~ number + gesttime + dose, data=litter)
[1] "Settings: unique SS : numeric variables centered"
> summary(mod2)
Component 1 :
Df R Sum Sq R Mean Sq Iter Pr(Prob)
number 1 64.85 64.849 4724 0.02075 *
gesttime 1 153.99 153.986 5000 0.00340 **
dose 3 122.80 40.934 5000 0.03720 *
Residuals 68 1086.42 15.977
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
There two things to note here. First, the aovp() function calculates unique sums of squares (also called Type III SS). Each effect is adjusted for all other effects, so order does not matter. Second, the dose effect is now significant (p = 0.038), suggesting that drug dose impacts birth weight after controlling for litter size and gestation period.
There are many situations were traditional linear model significance tests are not optimal (including when data is notably non-normal, there are outliers, and when sample sizes are too small to trust asymptotic results). In these cases, permuation tests may be viable alternatives.
To learn more about permuation tests in R see chapter 12 of R in Action.


One of the best sources that I used fir learning R is your web site,
thank you and good luck…
Thank you for the very informative blog. I have a question. I am using aovp() function to test for significant differences between BOLD signal properties in 3 groups adjusting for site and motion. I want to use a post-hoc test with the same model. I tried the TukeyHSD() function but this one does not work with 2 additional factors. Is there any post-hoc test you may suggest? Thanks in advance, Julia
Hi Julia,
I am not sure how I would design a post-hoc 3 group test with 2 covariates. It can get pretty complicated. I might approach this as an a-priori situation, compare the groups 2 at a time, controlling for the covariates, and then use the
p.adjust()function to control for alpha inflation.Hope this helps.
The lmPerm package is orphaned on 2014-02-11.
That is really a shame. I will have to transition to the coin package. Thanks for pointing this out.